






 |
 |
|||||||||||
| Function |  |
Structure |  |
Polymorphism |  |
Immunology |  |
Evolution |  |
Metabolism | ||
|
Function
Structure
Polymorphism
Immunology
Evolution
Metabolism
|
Access to the "Protein Structure Discovery" systemAccess to the "Protein Structure Discovery" system is authorized. Only users registered by the system administrator have the right to access its information. After the "Protein Structure Discovery" system is loaded, a window with a list of available operations appears on the screen (fig 1). 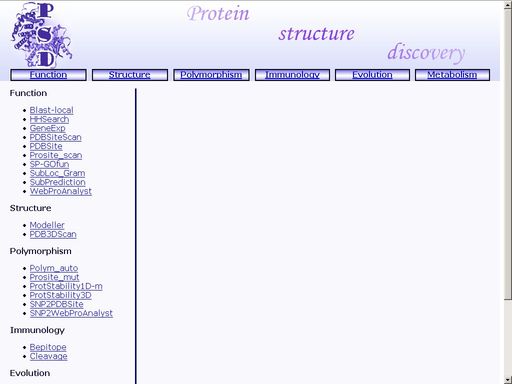
Figure 1. Interface of the "Protein Structure Discovery" system Click the preferred operation to start it. FunctionBlast-local - Search for homologs in protein annotated sequence databasesTo start the operation, enter the "Computer Proteomics" system and click "Blast-local". An HTML page with the web interface of the operation will appear, as shown in fig 2. 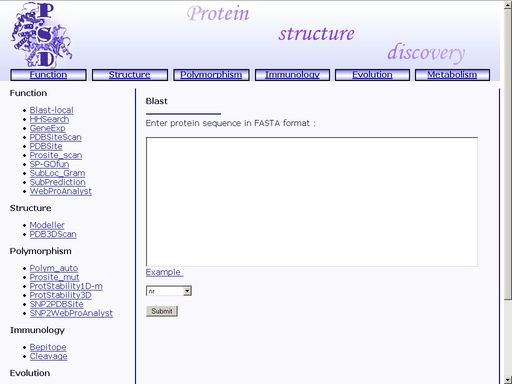
Figure 2. Interface of the "Blast-local" operation Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field, as shown in fig 3. 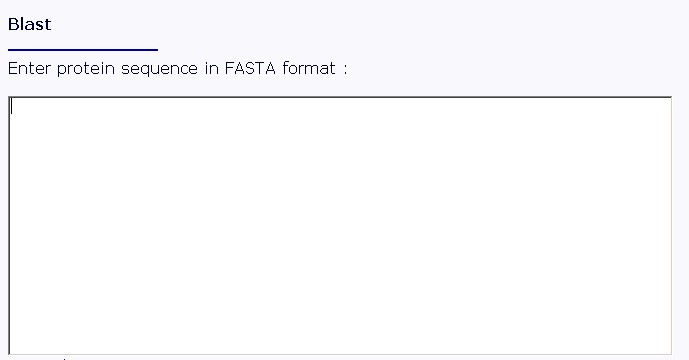
Figure 3. Text area for sequence input Also, you should specify a database to search against. You may choose it from the "Choose a database" list, as shown in fig 4. 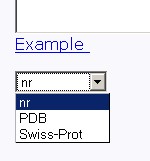
Figure 4. Choose a database Several databases are available: non-redundant (nr), Protein DataBase (PDB) and SwissProt (SwissProt). Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 4a. 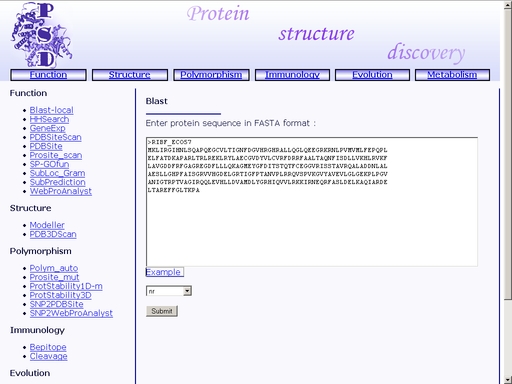
Figure 4a. A sample query Submit your query by clicking the "Submit" button below. When search is complete, you will get a HTML page with a result of your search. A sample result is shown in fig 5. 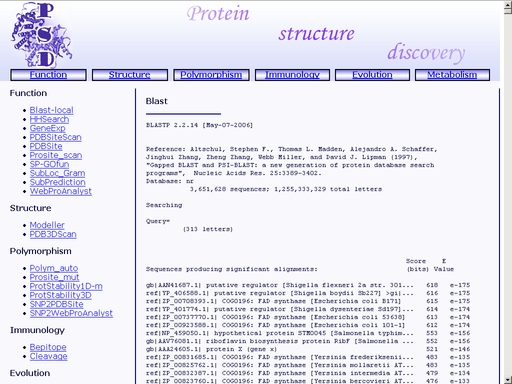
Figure 5. Result of the "Blast-local" operation You may save the result of your search as a text or html file using your web browser. Previously saved results may be used for further compative analysis. HHSearch - Search for homologs in protein functional family databases using Hidden Markov ModelsTo start the operation, enter the "Computer Proteomics" system and click "HHSearch". An HTML page with the web interface of the operation will appear, as shown in fig 2. 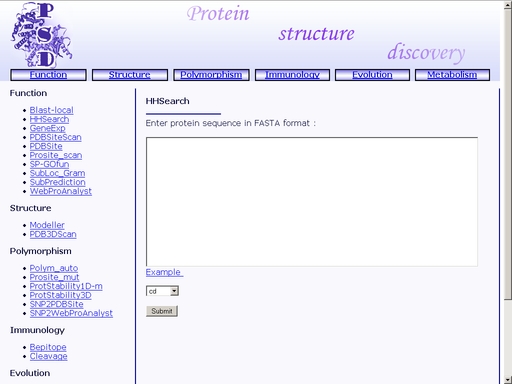
Figure 6. Interface of the "HHSearch" operation Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field, as shown in fig 7. 
Figure 7. Text area for sequence input Also, you should specify a database to search against. You may choose it from the "Choose a database" list, as shown in fig 8. 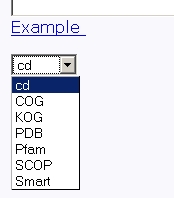
Figure 8. Choose a database Several databases are available: COG, PDB, Pfam, SCOP and Smart. Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 8a. 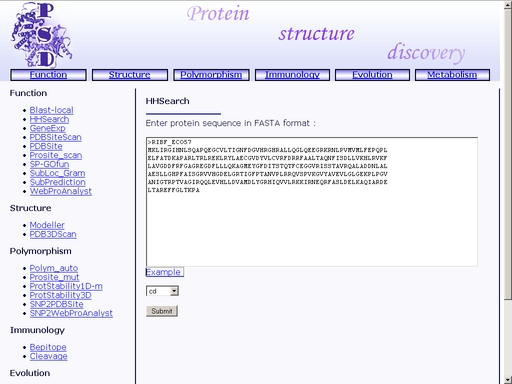
Figure 8a. A sample query Submit your query by clicking the "Submit" button below. When search is complete, you will get a HTML page with a result of your search. A sample result is shown in fig 9. 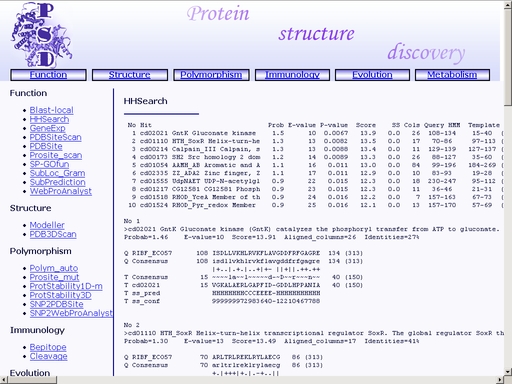
Figure 9. Result of the "HHSearch" operation You may save the result of your search as a text or html file using your web browser. Previously saved results may be used for further compative analysis. GeneExp - Prediction of gene expression levelTo start the operation, enter the "Computer Proteomics" system and click "GeneExp". An HTML page with the web interface of the operation will appear, as shown in fig 10. 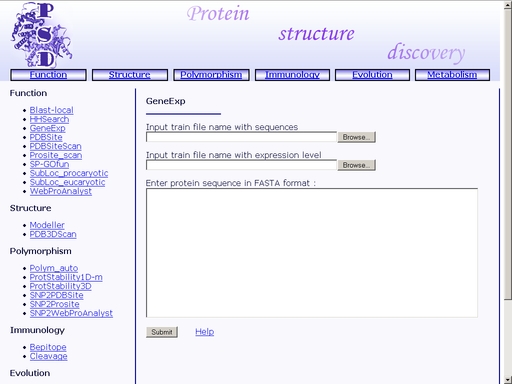
Figure 10. Interface of the "GeneExp" operation Upload amino acid sequences of proteins with known expression levels in FASTA format (text area "Input train file name with sequences"). These sequences will be used for learning. Click the "Browse" button to start standard upload dialog. Then upload a file with expression levels of the uploaded proteins (text area "Input train file name with expression level"). Click the "Browse" button to start standard upload dialog. Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field. Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 11. 
Figure 11. A sample query Submit your query by clicking the "Submit" button below. When prediction is complete, you will get an html page containing prediction of expression level of the gene that encodes the query protein. A sample result is shown in fig 12. 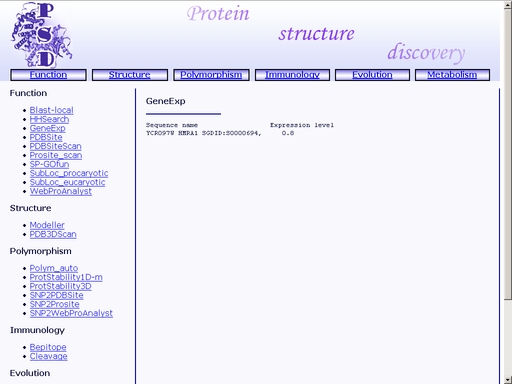
Figure 12. Result of the "GeneExp" operation PDBSiteScan - Recognition of functional sites in protein spatial structureTo start the operation, enter the "Computer Proteomics" system and click "PDBSiteScan". An HTML page with the web interface of the operation will appear, as shown in fig 2. 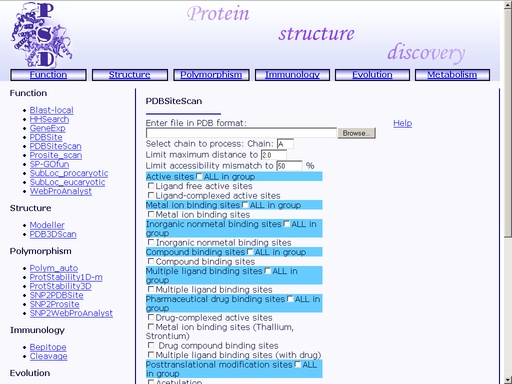
Figure 13. Initial state of the web interface of the "PDBSiteScan" operation. Meaning of the elements of the interface is as follows . 1) Text area for browsing a PDB file. 2) Text area for the chain ID input. 3) Input of MDM (Maximum Distance Mismatch) cutoff. 4) Cutoff for difference in solvent accessibility of site and template. 5) Checkbox for prediction of enzyme active sites. 6) Checkbox for prediction of metallic ion binding sites. 7) Checkbox for prediction of inorganic compound binding sites. 8) Checkbox for prediction of organic compound binding sites. 9) Checkbox for prediction of multiple ligand binding sites. 10) Checkbox for prediction of pharmaceutical compound binding sites. 11) Checkbox for prediction of sites of posttranslation modification in proteins. 12) Checkbox for prediction of sites of protein-protein interactions. 13) Checkbox for prediction of sites of protein-RNA interactions. 15) Checkbox for prediction of unclassified sites. To start prediction, upload a tertiary structure of your protein in PDB format. Click the "Browse" button to start standard upload dialog. This option fills the "Enter file in PDB format" text area with a path to your PDB file for you. Input one-letter identifier of the chain to be analyzed into the "Select chain to process" text area. By default, chain A will be analyzed. In addition, you may define MDM and solvent accessibility cutoffs (otherwise, the default values will be used). Also, you may search for functional sites of preferred types only, placing ticks in the corresponding checkboxes. Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 14. 
Figure 14. A sample query Submit your query by clicking the "Scan" button below. When prediction is complete, a web page with information on the found sites will appear, as shown in fig 15. 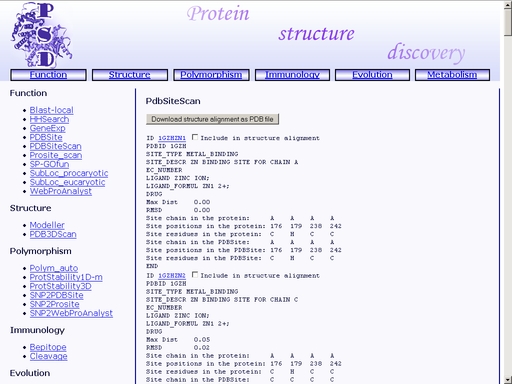
Figure 15. Result of the "PDBSiteScan" operation The format of the result page is as follows. 1) PDBSite template ID. Click the link to view the corresponding PDBSite entry. Place a tick in the checkbox "Include in structure alignment" to include the site in structure alignment of site templates with the query protein. 2) PDB identifier of the protein containing the site template. 5) Enzyme Classification number of the protein containing the site template 6) Ligand name. 7) Ligand chemical formula. 8) Pharmaceutical compound name (only if the ligand is a pharmaceutical compound). 9) Maximum Distance Match (MDM) and Root Mean Square Deviation (RMSD) values for the structural alignment of the site template with the query protein. 10) Chain identifier for the predicted site in the query protein. 11) Residue position numbers for the predicted site in the query protein. 12) Residue names in one-letter code for the predicted site in the query protein. 13) Chain identifier for the site template. 11) Residue position numbers for the site template. 14) Residue names in one-letter code for the site template. You may save your result for further analysis as a text file using your web browser. In addition, you may save structural alignment of the query protein with previously selected site templates in PDB format by clicking the "Download structure alignment as PDB file" button as shown in fig 16. This will open a standard dialog for a file save. 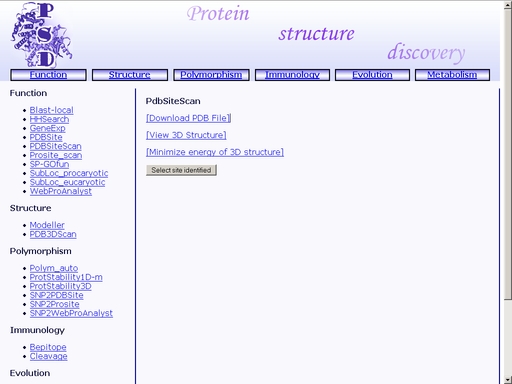
Figure 15. A file save dialog PDBSite - Database for protein functional site spatial structuresTo start the operation, enter the "Computer Proteomics" system and click "PDBSite". An HTML page with the web interface of the operation will appear, as shown in fig 16. 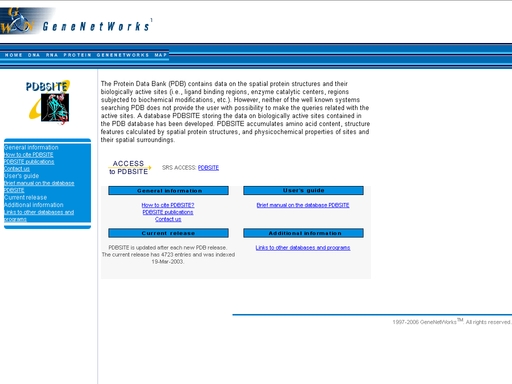
Figure 16. Initial state of the web interface of the "PDBSite" operation. Click the link "SRS ACCESS: PDBSITE" to start the SRS session for the PDBSite database, as shown in fig 17 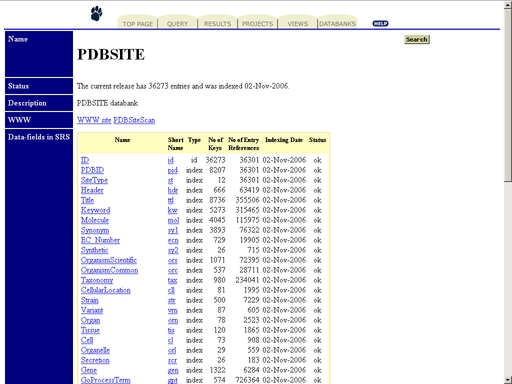
Figure 17. SRS interface of the PDBSite database. Prosite_scan - Search for motifs and patterns in protein primary structureTo start the operation, enter the "Computer Proteomics" system and click "Prosite_scan". An HTML page with the web interface of the operation will appear, as shown in fig 18. 
Figure 18. Interface of the "Prosite_scan" operation Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field, as shown in fig 19. 
Figure 19. Text area for sequence input Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 20. 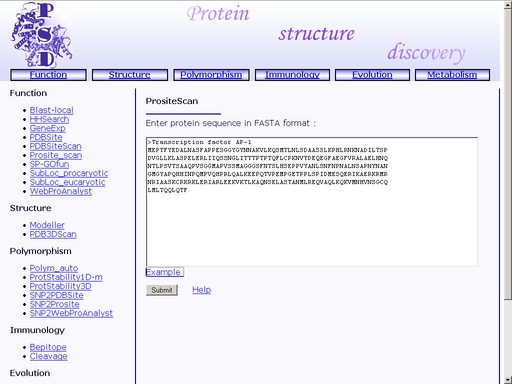
Figure 20. A sample query Submit your query by clicking the "Submit" button below. When search is complete, you will get a HTML page with a list of motifs and patterns found in the query sequence. A sample result is shown in fig 21. 
Figure 21. A sample html page with a result of search for motifs and patterns in the sample query. A. PROSITE ID of a pattern. B. Amino acid positions of the beginning and end of the pattern in the query sequence. C. The pattern sequence. You may save the result of your search as a text or html file using your web browser. The results may be used for further comparative analysis. SubLoc_procaryotic - Prediction of prokaryotic protein subcellular localizationTo start the operation, enter the "Computer Proteomics" system and click "SubLoc_prokaryotic". An HTML page with the web interface of the operation will appear, as shown in fig 2. 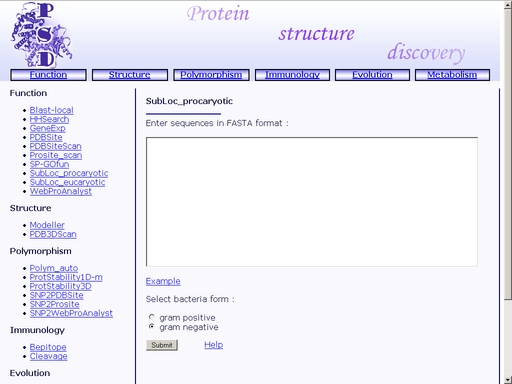
Figure 22. Interface of the "SubLoc_prokaryotic" operation Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field, as shown in fig 23. 
Figure 23. Text area for sequence input In addition, click one of the radio-buttons below, "gram-positive" or "gram-negative", to mark whether the bacteria, from which the query protein is extracted, is gram-positive or gram-negative, respectively. Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 24. 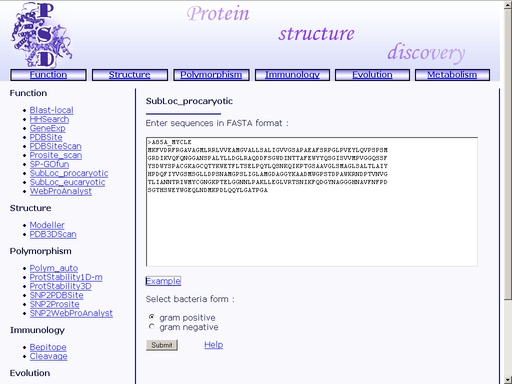
Figure 24. A sample query Submit your query by clicking "Submit" button below. You will get a web page with prediction of subcellular localization of the query protein, as shown in fig 25. 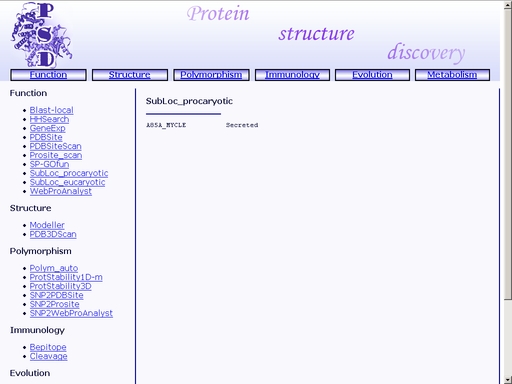
Figure 25. A sample result of the "SubLoc_prokaryotic" operation. SubLoc_eucaryotic - Prediction of eukaryotic protein subcellular localizationTo start the operation, enter the "Computer Proteomics" system and click "SubLoc_eukaryotic". An HTML page with the web interface of the operation will be brought up, as shown in fig 26. 
Figure 26. Interface of the "SubLoc_eukaryotic" operation Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field, as shown in fig 27. 
Figure 27. Text area for sequence input Once the sequence is entered, mark whether the query protein is extracted from animals, mushrooms, or plants. Click one of the radio buttons below to choose the kingdom the organism belongs to: "Animal" for animal species, "Fungi" for mushrooms and "Plant" for plants, as shown in fig 28. 
Figure 28. Choose the kingdom Submit your query by clicking the "Submit" button below. You will get a web page with prediction of the subcellular localization of the query protein (fig 29). 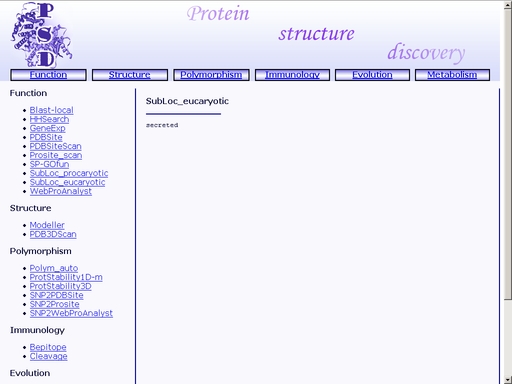
Figure 29. A sample result of the "SubLoc_eukaryotic" operation. WebProAnalyst - Prediction of Mutatons Having Directional Effect on Biological Activities and PropertiesTo start the operation, enter the "Computer Proteomics" system and click "WebProAnalyst". An HTML page with the web interface of the operation will appear, as shown in fig 30. 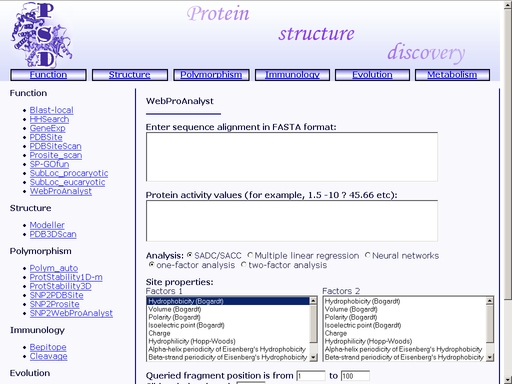
Figure 30. Interface of the "WebProAnalyst" operation Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field (see text area 1, fig 30). Additionally, enter a numeric activity value for each of the sequences. Enter "?" character to predict the activity of the corresponding protein (see text area 2, fig 30). Choose a statistical model, physicochemical properties of the protein sites, query fragment position in the alignment and input data type (see option menu 3, fig 30). Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 31. 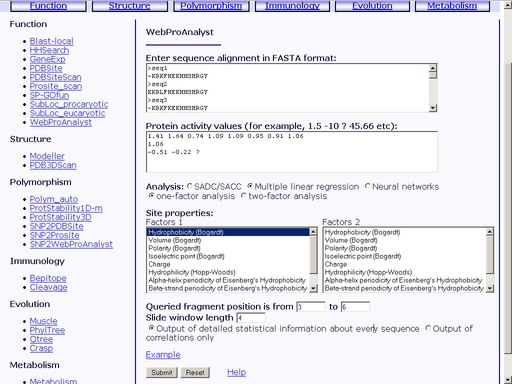
Figure 31. Meaning of the web interface elements is as follows. 1) Text area for input of multiple alignment. 2) Text area for input of activity values. 3) Option menu for selection of statistical model. 4, 5) Physicochemical properties to be used in statistical model of structure-activity relation. 6) Starting and ending positions of the alignment fragment to be analyzed. 7) Output data format (correlation values only or full statistical model). Submit your query by clicking the "Submit" button below. When prediction is complete, you will get an html page containing results on quantitative structure-activity relationship and prediction of activity of the query mutant protein. A sample result is shown in fig 31. 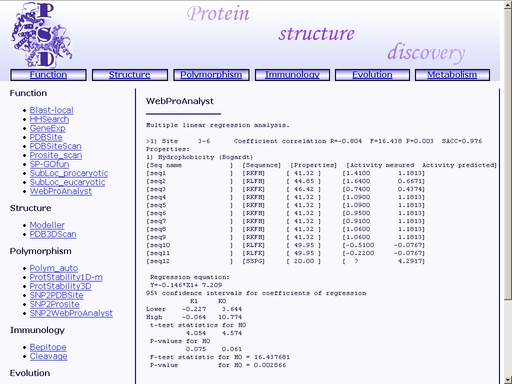
Figure 32. A sample result of the "WebProAnalyst" operation. SP-GOfun - Search for protein functionTo start the operation, enter the "Computer Proteomics" system and click "SP-GOfun". An HTML page with the web interface of the operation will appear, as shown in fig 33. 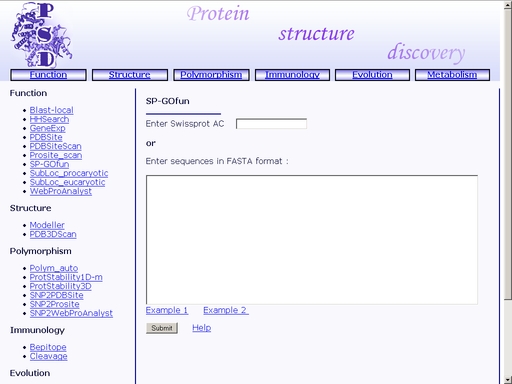
Figure 33. Interface of the "WebProAnalyst" operation Enter SwissProt Accession code of the query protein into the "Enter Swissprot AC" text area, as shown in fig 34. 
Figure 34. Text area for SwissProt Accession code input Also, you may enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field as shown in fig 35. 
Figure 35. Text area for SwissProt Accession code input Click the "Example 1" or "Example 2" link below to make a sample query. The query fields will automatically be filled for you, as shownt in fig 36a and 36b, respectively. 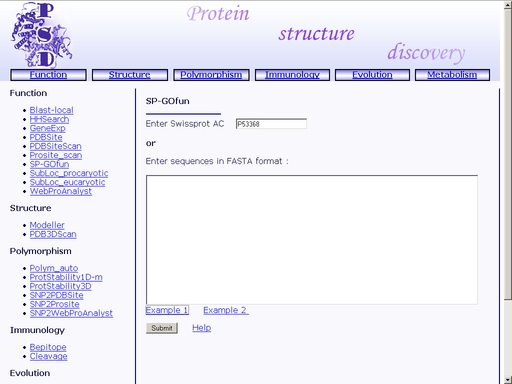
Figure 35a. A sample query 1. SwissProt Accession code input. 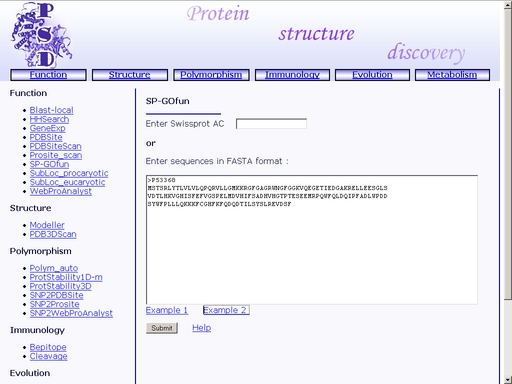
Figure 35a. A sample query 2. Amino acid sequence input. Submit your query by clicking the "Submit" button below. When prediction is complete, you will get an html page containing prediction of the query protein function. A sample result is shown in fig 36. 
Figure 36. A sample result of the "WebProAnalyst" operation. StructureModeller - Prediction of protein tertiary structureTo start the operation, enter the "Computer Proteomics" system and click "Modeller". An HTML page with the web interface of the operation will appear, as shown in fig 37. Figure 37. Interface of the "Modeller" operation Enter the identifier of the template protein tertiary structure and its polypeptide chain in Protein DataBase (PDB) into the appropriate text areas: "Enter PDB id" and "Enter chain id". You may leave the areas blank, then Modeller will search its own database for the best template. Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field. Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 38. Figure 38. A sample query. Submit your query by clicking the "Submit" button below. When prediction is complete, a standard dialog for a file save will be opened automatically. You will offered to save prediction of the query protein tertiary structure. A sample result as a file in PDB format. A sample dialog is shown in fig 39. Figure 39. A file save dialog PDB3DScan - Search for structural homologs in PDB database using structural alignmentTo start the operation, enter the "Computer Proteomics" system and click "PDB3DScan". An HTML page with the web interface of the operation will appear, as shown in fig 40. Figure 40. Interface of the "PDB3DScan" operation Upload a tertiary structure of your protein in PDB format. Click the "Browse" button to start standard upload dialog. This option fills the "Input PDB filename" text area with a path to your PDB file for you. Input one-letter identifier of the chain to be analyzed into the "Enter chain" text area, as shown in fig 41. By default, chain A will be used. Figure 41. Structure upload Submit your query by clicking the "Submit" button below. When search is complete, you will get an html page with a list of structural homologs of the query protein. A sample result is shown in fig 42. Figure 42. A sample result of the "WebProAnalyst" operation. PolymorphismPolym_auto - Database for human gene and protein polymorphismsTo start the operation, enter the "Computer Proteomics" system and click "Polym_auto". This will start the SRS session for the Polym_auto database, as shown in fig 43. Figure 43. SRS interface of the Polym_auto database. ProtStability1D-m - Prediction of mutation effect on protein thermodynamic stability based on primary structureTo start the operation, enter the "Computer Proteomics" system and click "ProtStability1D-m". An HTML page with the web interface of the operation will appear, as shown in fig 44. Figure 44. Interface of the "ProtStability1D-m" operation Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequence in FASTA format" field. Optionally, you may analyze a certain substitution by entering position and amino acid type for your mutation into the text areas "Mutation position" and "Mutation amino acid type", respectively. Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 45. Figure 45. A sample query. Submit your query by clicking the "Submit" button below. When prediction is complete, you will get an html page with prediction of mutational effect on thermodynamic stability of the query protein, as shown in fig. 46. Figure 46. A sample result of the "ProtStability1D-m" operation. SNP2PDBSite - Prediction of Mutational Effect on Modification of Functional Sites, Their Appearance and DeletionTo start the operation, enter the "Computer Proteomics" system and click "SNP2PDBSite". An HTML page with the web interface of the operation will appear, as shown in fig 47. Figure 47. Interface of the "SNP2PDBSite" operation Enter the identifier of the query protein tertiary structure and its polypeptide chain in the Protein DataBase (PDB) into corresponding text areas: "Enter PDB id" and "Enter chain id". Also, enter a list of amino acid substitutions to be analyzed. The format for a substitution entry is as follows: chain_id residue_number wild_type_residue -> mutant_residue For example, enter A 245 G -> C for G245C substitution in chain A. Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 48. Figure 48. A sample query. Submit your query by clicking the "Submit" button below. When prediction is complete, you will get an html page with a list of arisen, altered, or deleted sites, as shown in fig 49. Figure 49. A sample result of the "SNP2PDBSite" operation. SNP2WebProAnalyst - Prediction of mutant protein activityTo start the operation, enter the "Computer Proteomics" system and click "SNP2WebProAnalyst". An HTML page with the web interface of the operation will appear, as shown in fig 50. Figure 50. Interface of the "SNP2WebProAnalyst" operation Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field as shown in fig 51. Figure 51. Text area for sequence input Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 52. Figure 52. A sample query Submit your query by clicking the "Submit" button below. When prediction is complete, you will get an html page containing results on quantitative structure-activity relationship and prediction of activity of the query mutant protein. A sample result is shown in fig 31. Figure 53. A sample result of the "SNP2WebProAnalyst" operation. SNP2Prosite - Prediction of Mutational Effect on Modification of Amino Acid Sequences of Functional Sites, Their Appearance and DeletionTo start the operation, enter the "Computer Proteomics" system and click "SNP2Prosite". An HTML page with the web interface of the operation will appear, as shown in fig 54. Figure 54. Interface of the "SNP2Prosite" operation Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field, as shown in fig 3. Then enter a list of amino acid substitutions to be analyzed. The format for a substitution entry is as follows: chain_id residue_number wild_type_residue -> mutant_residue For example, enter A 245 G -> C for G245C substitution in chain A. Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 55. Figure 55. A sample query Submit your query by clicking the "Submit" button below. When prediction is complete, you will get an html page with a list of arisen, altered, or deleted sites, as shown in fig 56. Figure 56. A sample result of the "SNP2WebProAnalyst" operation. ProtStability3D - Prediction of Mutational Effect on Thermodynamical Stability of a Protein Using its Tertiary StructureTo start the operation, enter the "Computer Proteomics" system and click "ProtStability3D". An HTML page with the web interface of the operation will appear, as shown in fig 57. Figure 57. Interface of the "ProtStability3D" operation Enter the identifier of the query protein tertiary structure and its polypeptide chain in Protein DataBase (PDB) into the appropriate text areas: "Enter PDB id" and "Enter chain id". Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 58. Figure 58. A sample query Click the "Submit" button below to submit your query. When prediction is complete, you will get an html page with prediction of mutational effect on thermodynamic stability of the query protein, as shown in fig. 59 Figure 59. A sample result of the "ProtStability3D" operation. ImmunologyCleavage - Proteosomal Cleavage Sites PredictionTo start the operation, enter the "Computer Proteomics" system and click "Cleavage". An HTML page with the web interface of the operation will appear, as shown in fig 60. Figure 60. Interface of the "Cleavage" operation Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field as shown in fig 61. Figure 61. Text area for sequence input Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 62. Figure 62. A sample query Submit your query by clicking the "Submit" button below. When prediction is complete, you will get an html page with a list of cleavage sites of the query protein. A sample result is shown in fig 63. Figure 63. A sample result of the "Cleavage" operation. Bepitope - B-epitope predictionTo start the operation, enter the "Computer Proteomics" system and click "Bepitope". An HTML page with the web interface of the operation will appear, as shown in fig 64. Figure 64. Interface of the "Bepitope" operation Enter the amino acid sequence of your query protein. Query protein sequence in one-letter code {A, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y, a, c, d, e, f, g, h, i, k, l, m, n, p, q, r, s, t, v, w, y} must not be shorter than 25 residues and not longer than 2000 residues in FASTA format. You may type or paste the sequence into the "Enter sequences in FASTA format" field, as shown in fig 65. Figure 65. Text area for sequence input Click the "Example" link below to make a sample query. The query fields will automatically be filled for you, as shown in fig 66. Figure 66. A sample query Submit your query by clicking the "Submit" button below. When prediction is complete, you will get an html page with a list of B-epitopes of the query protein. A sample result is shown in fig 67. Figure 67. A sample result of the "Bepitope" operation. |
 |
|
| ICG©2006-2016 |
Designed by EVA |