About
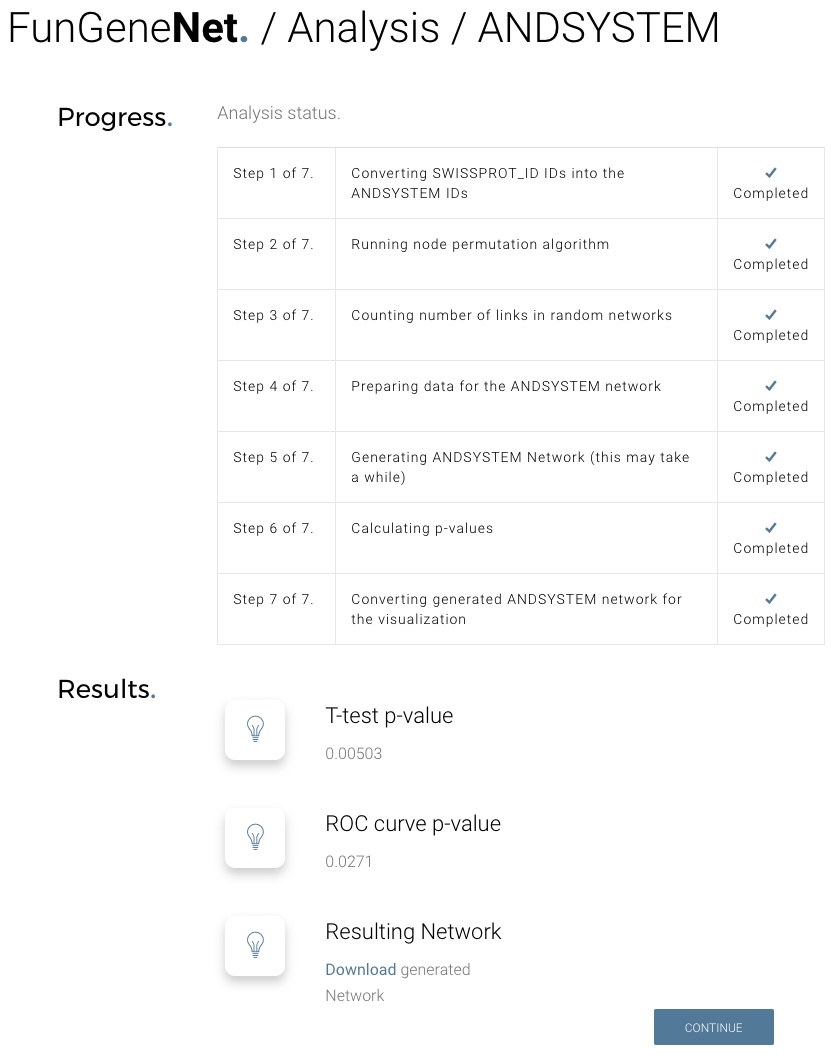
FunGeneNet is a web tool aimed at proving the functionality of networks in a wide range of sizes of experimental gene sets, both for different global networks and for different types of interactions. It utilizes the ANDSystem and STRING to reconstruct gene networks using experimental gene sets, provided by the user, and to estimate their difference from random networks. A main feature of the method implemented in FunGeneNet is that it allows consideration of specific types of molecular-genetics interactions, while most of the existing methods only work with generalized types characterizing the existence of any interaction between network members. The sensitivity of our method exceeds 0.8, the specificity is 0.95.